There are 10-100 trillion microbes that reside in our gastrointestinal tract, representing thousands of species. The gut microbiota is a control center for multiple aspects of our biology including our immune status, metabolism, and neurobiology. Many of the metabolic activities and developmental signals that the microbiota deploys are complementary to our own, affirming that humans are composite organisms consisting of microbial and human parts.
Microbiota composition varies considerably between individuals, and factors like variation in host genotype and diet impact the community. Changes in our microbial communities differentially influence aspects of host biology (e.g., immune function) and likely explain aspects of variation within and between human populations (e.g., predisposition to disease). The plasticity of the microbiota suggests that it may be a viable therapeutic target, and necessitates the pursuit of a fundamental understanding of how extrinsic and intrinsic factors alter its composition, function, and interaction with the host.
Our research program aims to elucidate the basic principles that govern interactions within the intestinal microbiota and between the microbiota and the host. Specifically, we are exploring the effects of perturbations in the intestinal environment, such as changes in diet, microbial community composition, pathogen exposure, host genotype, and microbiota-targeted small molecules. To pursue these aims, we study germ-free mice colonized with simplified, model microbial communities, apply systems approaches and use genetic tools for the host and microbes to gain mechanistic insight into emergent properties of the host-microbial superorganism.
Diet is a major determinant of which microbes flourish in the microbiota. Dietary microbiota-accessible carbohydrates (MACs), the main component of dietary fiber, serve as the primary metabolic input for the gut microbiota. However, during times of dietary MAC scarcity, the microbiota turn to host mucus for sustenance. We are interested in the mechanisms that underlie microbiota consumption of dietary MACs, and how these components of our diet dictate microbiota composition, function, and diversity to influence host biology.
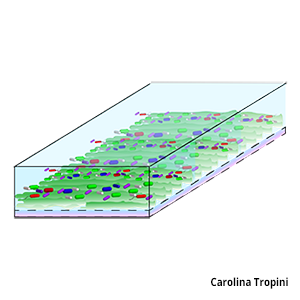
We have learned much about the microbiota via survey/sequencing bulk fecal material, however within the gastrointestinal tract there exist a number of distinct microenvironments: the colonic crypts, the loose outer mucus layer, the lumen, and the length of the gut from small intestine to descending colon. Variations in pH, carbon sources, host immune effectors and small molecules likely shape local community composition and spatial dynamics, which in turn can impact gastrointestinal health. We are using quantitative imaging to define how host diet, genetics and environment affect the localization of commensal microbes in both healthy and perturbed states.

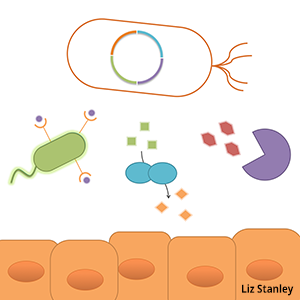
In a healthy and complex microbiota, space and resources are occupied by commensal species, providing colonization resistance to potential pathogens that may pass through, or reside within, the gut. However, when perturbations like antibiotics or diarrhea disrupt the resident microbes, colonization resistance and efficient nutrient networks are compromised. Pathogens like Salmonella typhimurium and Clostridium difficile are able to exploit temporary availability of resources and expand. Our lab seeks to understand the mechanisms by which the gut microbiota protects its host from gastrointestinal infections and how interactions between the microbiota, pathogens, and host steer the course of infection.
The intestinal microbiota continually produces numerous small molecules that may be absorbed into circulation and represent a dynamic and ill- defined contribution to host biology and metabolism. Despite the vast metabolic capabilities encoded by the human microbiome, little is known about the species and genes responsible for producing such molecules or how their production may be manipulated, for instance by diet or by microbiota transplant. We are mapping the relationships between diet, microbial metabolic pathways, and the most abundant and biologically influential drug-like compounds manufactured by the microbiota.
Gut bacteria possess a repertoire of metabolic strategies, defense mechanisms and environmental sensors highly adapted for thriving in the gut environment, suggesting these microbes can be considered a technology platform for influencing human health. The ability to record the environmental experiences, rewire behavior, or add new functions to these commensal microbes will help us understand how they respond to and influence their environment, and provide a means for engineering bacteria-delivered therapeutics in the future. High-throughput strain modification techniques, tunable protein expression, protein secretion and logic and memory devices are key technologies we are developing to enable a wide range of studies and applications.
The gut microbiota has to respond very quickly to physical changes happening in the gut. Current methods of inquiry that rely on sequencing or tissue imaging are not able to probe both temporal and spatial resolution at fine scales. We are therefore developing microfluidic devices to culture the gut microbiota in microfluidic mucosal layers. The ability to monitor the real-time dynamics of a complex microbial community during physical perturbations will allow us to characterize relevant members and key metabolic pathways involved during disruption of the normal equilibrium. To achieve fine-scale microfluidic control we use an Elveflow controller.